5 Open science
5.1 Learning objectives
In this chapter, you will learn the reasons for practicing open science and some of the basic methodological techniques that we can use to facilitate an open science workflow.
5.2 What is Open Science and why?
Open Science is a way of doing research that emphasizes public and free accessibility, reusability, and transparency at every stage of the scientific process. Instead of keeping data, methods, and results behind paywalls or locked on personal computers, Open Science seeks to make them available for others to examine, test, and build upon. In brief:
“Open data and content can be freely used, modified, and shared by anyone for any purpose.” (opendefinition.org)
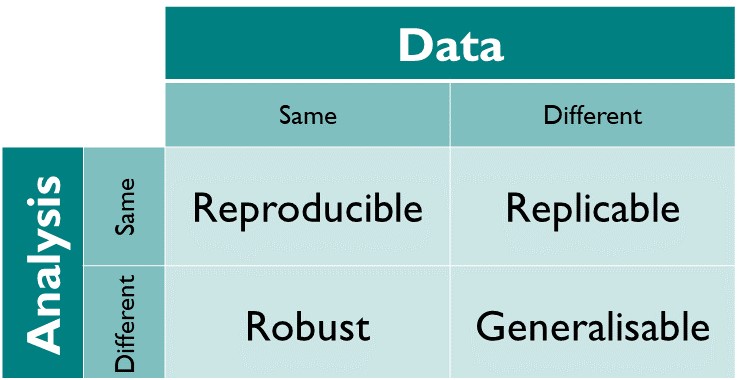
At its core, Open Science is motivated by the requirement for reproducibility of scientific findings: Results should be testable by others using the same methods and data. More fundamentally, results should be replicable, robust, and generalisable. These descriptors can be understood as different combinations of the data and/or the analysis being identical between studies (or not). Open access data and software are the basis for a study to be reproducible. The analysis (software) may be reusable and can be applied to new data, making findings replicable. The same data may also be analysed differently. If findings are similar, they are robust. If a pattern or a relationship is identified consistently with multiple different combinations of data and analytic approaches, findings are generalisable. Within the context of this course, we focus primarily on the practical aspects for reproducible science, i.e., ensuring that given the same data and code, the results will be identical.
Reusability of code in project-oriented workflows (see Chapter 3) is key to Open science. It ensures that outputs such as datasets, software, and workflows can be adapted for new purposes, speeding up discovery across disciplines. Concretely, Open Science includes practices including open data, open source software, open access publishing, and open workflows and methods (see below). Beyond methods, Open Science represents a cultural shift. It challenges the traditional “publish or perish” system, where competition and secrecy often slow progress, and instead encourages collaboration, accountability, and broader participation. Several aspects motivate the cultural shift towards Open Science:
- Enables accountability and trust, and leads to robust science.
Access to data and code enables tests and acts as a precise documentation of how results were obtained. This also reflects a response to the reproducibility crisis that has affected scientific studies and entire scientific fields.
- Creates efficient infrastructure.
Making data freely accessible through the web enables fully reproducible workflows and the integration of data access into reproducible and continuously updated workflows.
- Enhances impact.
Making your data and code accessible and reusable enhances its uptake by the community. This makes your work valuable and visible – you create impact.
- Serves the public and transcends discrimination.
Since much research is publicly funded, its outputs should be accessible to the public and anyone, irrespective of access to resources and funding.
5.2.1 Open data
Open data refers to the practice of making research datasets available so that others can examine, validate, and reuse them. When properly shared, data becomes a resource for the entire research community, enabling new insights and reducing duplication of effort. Yet, in practice, researchers often perceive barriers to making data open. In some cases, researchers must handle sensitive or confidential data, which cannot be openly released without consent and restrictions to openness. Finally, there are strategic considerations—sharing data may feel risky in competitive environments.
Despite these challenges, expectations and the culture around open data are shifting. Many journals and funding agencies now require authors to provide data availability statements in published articles, and some make data deposition in recognized open research data repositories a condition of publication. To guide these practices, the FAIR principles (Wilkinson et al. 2016) – that data should be Findable, Accessible, Interoperable, and Reusable – have become a global standard. FAIR does not mean all data must be open, but it emphasizes that well-documented and properly archived data maximizes its potential for reuse while protecting sensitive information.
To ensure long-term storage of code and data, outside of commercial for profit services (e.g., Dropbox, Google Drive etc), it is best to rely on public permanent repositories, such as Zenodo. Zenodo is an effort by the European commission, but accessible to all, to facilitate archiving of science projects of all nature (code and data) up to 50 GB. In addition, Zenodo provides a citable digital object identifier or DOI. This allows data and code, even if not formally published in a journal, to be cited. Other noteworthy open science storage options include Dryad and the Center for Open Science.
The broad-purpose permanent data repositories mentioned above are not edited and are therefore not ideal for data discovery. In contrast, edited data repositories often have a specific thematic scope and different repositories are established in different research communities. Table 5.1 provides a list of widely used data repositories, generalist and others, that provide manual or automated download access to their data. Note that this list contains some example and is far from extensive.
| Repository name | URL | Purpose | Open to contributions |
|---|---|---|---|
| Zenodo | https://zenodo.org/ | Generalist data repository for research data. | Yes |
| Figshare | https://figshare.com/ | Generalist data repository for research data. | Yes |
| Dryad | https://datadryad.org | Generalist data repository for research data. | Yes |
| Environmental Data Initiative | https://edirepository.org/ | Hosts ecological and environmental data | Yes |
| World Data Center for Climate | https://www.wdc-climate.de/ui/ | Hosts Earth system model data, run by Deutsches Klimarechenzentrum (DKRZ) | Yes |
| PANGAEA | https://www.pangaea.de/ | Hosts georeferenced data from earth system research | Yes |
| EnviDat | https://www.envidat.ch | Hosts envrionmental system research with a focus on Switzerland | For researchers of the Swiss Federal Institute for Forest, Snow and Landscape WSL |
| Climate Data Store | https://cds.climate.copernicus.eu/ | Provides access to climate data | No |
| Copernicus Data Space Ecosystem | https://dataspace.copernicus.eu/ | Provides access to data from the Copernicus Sentinel missions | No |
| Meteoswiss Open Data | https://opendatadocs.meteoswiss.ch/ | Switzerland-specific meteorological data | No |
| Swiss Open Government data | https://opendata.swiss/en/ | Generalist data from the Swiss Federal statistics office. | No |
| Eurostat | https://ec.europa.eu/eurostat | Provides access to statistical data of Europe | No |
5.2.2 Open source software
Software is often as central to research as data, yet historically it has been less visible in scholarly outputs. Open source software addresses this gap by making code freely accessible for anyone to use, examine, and extend. A typical project might start as a tool written for a specific study, but once shared and adapted for more general applications, it can evolve into a library that supports many projects across a community. Also specialized software, such as a model for simulating a physical system, can become widely useful if written in a non-proprietary coding language and accompanied by sufficient documentation.
The benefits of open source are significant. Publicly available code enables reproducibility by letting others see exactly how analyses were performed. It also reduces dependence on proprietary systems, which may lock research into costly or opaque environments. Over time, well-maintained open source projects can attract contributions from the wider community, enhancing functionality and improving reliability. Version control platforms like GitHub make this collaborative development possible by offering tools for issue tracking, pull requests, and continuous integration testing. When paired with clear licensing, open source software not only supports transparency but also lays the foundation fair crediting and for cumulative progress.
A challenge in sopen source software lies in standardisation, compatibility, and stability. Libraries hosted on central repositories that maintain tests and standards on contributed software are essential to making an open software environment work. For R, CRAN is the central library repository. However, not all libraries, developed for specific science applications may be suitable for CRAN as it enforces certain standards that may be too restrictive. ROpenSci is an alternative for well-documented open research software.
5.2.3 Open access publishing
One of the most visible aspects of Open Science is open access publishing, which aims to make research articles freely available to everyone. The motivation is straightforward: since much research is publicly funded, the public should not face paywalls to access it. However, the reality of traditional publishing has created a crisis. Commercial publishers often charge universities and libraries high subscription fees, while researchers – who generate content, review manuscripts, and edit journals without pay – receive little in return. The public ends up paying twice: once to fund the research through salaries and grants, and again through subscription costs.
Open access offers alternative models. Some scholarly societies, such as the European Geosciences Union and the American Geophysical Union, have pioneered non-profit publishing routes where revenues sustain the scientific community rather than shareholders. Preprint servers also play an increasingly important role, allowing researchers to freely share findings rapidly before peer review.
Within formal publishing, two main pathways to open access exist. Gold open access makes articles immediately free to read on the publisher’s site, often supported by article processing charges (APCs) paid by authors or their institutions. Green open access relies on authors self-archiving preprints or accepted manuscripts in institutional or subject repositories, where they remain freely available even if the journal version is behind a paywall. Together, these models are reshaping the landscape of scholarly communication toward greater openness.
5.2.4 Open workflows and methods
Beyond data, software, and publications, Open Science also encompasses the entire research workflow. Open workflows mean that the processes behind a study – from data collection protocols to analysis scripts – are documented and shared in ways that others can follow. This makes research more transparent and easier to reuse. For example, maintaining project-oriented workflows where data, code, and documentation are stored systematically helps others (and one’s future self) understand the work. In Chapter 3, you learned what it takes to implement reusable, project-oriented workflows.
Version control systems like Git allow researchers to capture the history of their analysis workflows, while platforms like GitHub provide mechanisms for collaboration, peer review of code, and continuous integration testing to ensure reliability. Coupled with practices like versioning and tagged releases, these tools enable researchers to share stable snapshots of their work that can be cited and archived. Open workflows are also supported by data management plans, which assign clear roles and responsibilities, define storage and backup strategies, and address long-term preservation of outputs. More on such practices below in Section 5.3
In essence, open workflows transform research into a living, transparent process rather than a black box that only produces final results. This shift helps ensure that scientific claims can be checked, methods can be reused, and future work can build more efficiently on what has already been done.
5.3 Practices for Open Science
5.3.1 Open data
Practices for applying FAIR data principles involve multiple methods and resources, relevant for different steps of data generation and publication.
Organise and clean your data
- Remove duplicates, errors, or inconsistencies.
- Use clear, descriptive variable names (column names in tabular data).
- Standardize units of measurement.
- Make contents machine-readable (see Section 7.3.10)
Choose an open, non-proprietary format
- For tabular data, use CSV.
- For structured scientific data, consider formats like NetCDF or HDF5, which support embedded metadata.
- For text, use txt or Markdown rather than proprietary formats (e.g., DOCX).
- For images or media, use open formats like PNG, TIFF, WAV, or MP4.
Document your dataset
- Create a README file (plain text or Markdown, see also Section 3.4.3.1) that contains the following information:
- Dataset title and description.
- Explanation of variables (column names, units, categories).
- Methods used for data collection and processing.
- Links to related publications, software, or workflows.
- Author names, affiliations, and contact information.
- License information (e.g., Creative Commons).
Deposit your data in a trusted open repository
- Use a permanent repository with strong archival policies (see Table 5.1).
- Ensure the repository generates a digital object identifier (DOI) for long-term citation of uploaded datasets
- Add metadata to make the dataset discoverable (keywords, subject categories, abstract).
Integrate data into a repository or package
- Data may be contained in a Git repository and published through that. This only makes sense if the data is stored in text-based files (e.g., CSV) and is permissible only for data files of limited size (<25 MB on Github).
- This allows clear versioning through a tag and release and allows tracking the state of the data through the commit history.
- Link your Github repository to Zenodo for automatic DOI assignment when releasing a version (see here and here).
- Extend the repository to make in library (Example: leaf13C).
Provide a data availability statement
- Clearly indicate in your publications where the data can be found.
- State the license and any access restrictions. Use available licenses, see e.g., Creative Commons.
- If data is sensitive and cannot be shared, provide metadata and conditions for access.
5.3.2 Open source software
Developing open source software for research is more than simply sharing code – it involves adopting practices that make your code reusable, sustainable, and valuable to the broader community. The following tools and methods are crucial to making code open.
Make it reusable
- Adopt a reusable project-oriented workflow (see Chapter 3).
Use version control from the start
- Make the project directory a Git repository from the start, hosted, e.g., on Github.
Make stable releases
- Use a git tag to mark major updates of the repository. These may align with designated version names (e.g.,
v1.0.0). - Follow semantic versioning – a standardised way of using version numbering to designate major, minor, and patch changes to the code.
- Create releases of tags on GitHub or GitLab with notes describing changes.
- Link your Github repository to Zenodo for automatic DOI assignment when releasing a version (see here and here).
Refactor reusable components into a library
- Further enhance reusability of functions that may be re-used across projects by creating a library (package).
- Use a non-proprietary coding language (e.g., R, Python, Julia) for maximum accessibility of the package.
- See here for a comprehensive tutorial on writing R packages.
Document
- Write vignettes (see Section 3.4.3.4) or tutorials showing use cases.
- Create a project website with documentation (e.g., using GitHub Pages, and pkgdown for R).
- Publish a software paper in open software description journal (e.g., Journal of Open Source Software).
- Submit your library to a central archive: CRAN for R, or PyPI for Python.
Maintain and improve the project
- Keep track of issues for bug reports and feature requests.
- Keep a changelog of updates and fixes.
- Encourage community contributions through pull requests and clear contributing guidelines.