Code
library(dplyr)
library(readr)
library(purrr)
library(ggplot2)
library(terra)
library(tidyterra)
library(leaflet)
library(sf)
library(here)
library(knitr)The data required for ditigal soil mapping are:
Let’s load them
| site_id_unique | timeset | x | y | dataset | dclass | waterlog.30 | waterlog.50 | waterlog.100 | ph.0.10 | ph.10.30 | ph.30.50 | ph.50.100 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4_26-In-005 | d1968_1974_ptf | 2571994 | 1203001 | validation | poor | 0 | 0 | 1 | 6.071733 | 6.227780 | 7.109235 | 7.214589 |
| 4_26-In-006 | d1974_1978 | 2572149 | 1202965 | calibration | poor | 0 | 1 | 1 | 6.900000 | 6.947128 | 7.203502 | 7.700000 |
| 4_26-In-012 | d1974_1978 | 2572937 | 1203693 | calibration | moderate | 0 | 1 | 1 | 6.200000 | 6.147128 | 5.603502 | 5.904355 |
| 4_26-In-014 | d1974_1978 | 2573374 | 1203710 | validation | well | 0 | 0 | 0 | 6.600000 | 6.754607 | 7.200000 | 7.151129 |
| 4_26-In-015 | d1968_1974_ptf | 2573553 | 1203935 | validation | moderate | 0 | 0 | 1 | 6.272715 | 6.272715 | 6.718392 | 7.269008 |
| 4_26-In-016 | d1968_1974_ptf | 2573310 | 1204328 | calibration | poor | 0 | 0 | 1 | 6.272715 | 6.160700 | 5.559031 | 5.161655 |
The dataset on soil samples from Bern holds 13 variables for 1052 entries (more information here):
site_id_unique: The location’s unique site id.
timeset: The sampling year and information on sampling type for soil pH (no label: CaCl\(_2\) laboratory measurement, field: indicator solution used in field, ptf: H\(_2\)O laboratory measurement transferred by pedotransfer function).
x: The x (easting) coordinates in meters following the (CH1903/LV03) system.
y: The y (northing) coordinates in meters following the (CH1903/LV03) system.
dataset: Specification whether a sample is used for model training ("calibration") or testing ("validation") (this is based on randomization to ensure even spatial coverage).
dclass: Soil drainage class
waterlog.30, waterlog.50, waterlog.100: Specification whether soil was water logged at 30, 50, or 100 cm depth (0 = No, 1 = Yes).
ph.0.10, ph.10.30, ph.30.50, ph.50.100: Average soil pH between 0-10, 10-30, 30-50, and 50-100 cm depth.
Now, let’s load the covariates that we want to produce our soil maps with. These files are in the geoTIFF format - geolocated TIFF files.
[[1]]
[1] "NegO.tif"
[[2]]
[1] "PosO.tif"
[[3]]
[1] "Se_MRRTF2m.tif"
[[4]]
[1] "Se_MRVBF2m.tif"
[[5]]
[1] "Se_NO2m_r500.tif"
[[6]]
[1] "Se_PO2m_r500.tif"The output above shows the first five raster files with rather cryptic names. The meaning of all 91 raster files are given in Chapter 6. Make sure to have a look at that list as it will help you to interpret your model results later on. Let’s look at one of these raster files, Se_slope2m.tif, to get a better understanding for our data. That file contains the local slope of the terrain, derived from a digital elevation model with 2 m resolution:
class : SpatRaster
size : 986, 2428, 1 (nrow, ncol, nlyr)
resolution : 20, 20 (x, y)
extent : 2568140, 2616700, 1200740, 1220460 (xmin, xmax, ymin, ymax)
coord. ref. : CH1903+ / LV95
source : Se_slope2m.tif
name : Se_slope2m
min value : 0.00000
max value : 85.11286 As shown in the output, a raster object has the following properties (among others, see ?rast):
class: The class of the file, here a SpatRaster.
dimensions: The number of rows, columns, years (if temporal encoding).
resolution: The resolution of the coordinate system, here it is 20 in both axes.
extent: The extent of the coordinate system defined by min and max values on the x and y axes.
coord. ref.: Reference coordinate system. Here, the raster is encoded using the LV95 geodetic reference system from which the projected coordinate system CH1903+ is derived.
source: The name of the source file.
names: The name of the raster file (mostly the file name without file-specific ending)
min value: The lowest value of all cells.
max value: The highest value of all cells.
The code chunks filtered for a random sub-sample of 15 variables. As described in Chapter 5, your task will be to investigate all covariates and find the ones that can best be used for your modelling task.
Now, let’s look at a visualisation of this raster file. Since we have selected the slope at 2 m resolution, we expect a relief-like map with a color gradient that indicates the steepness of the terrain. A quick way to look at a raster object is to use the generic plot() function.
To have more flexibility with visualising the data, we can use the ggplot() in combination with the {tidyterra} package.
# To have some more flexibility, we can plot this in the ggplot-style as such:
ggplot() +
tidyterra::geom_spatraster(data = raster_example) +
scale_fill_viridis_c(
na.value = NA,
option = "magma",
name = "Slope (%) \n"
) +
theme_bw() +
scale_x_continuous(expand = c(0, 0)) + # avoid gap between plotting area and axis
scale_y_continuous(expand = c(0, 0)) +
labs(title = "Slope of the Study Area")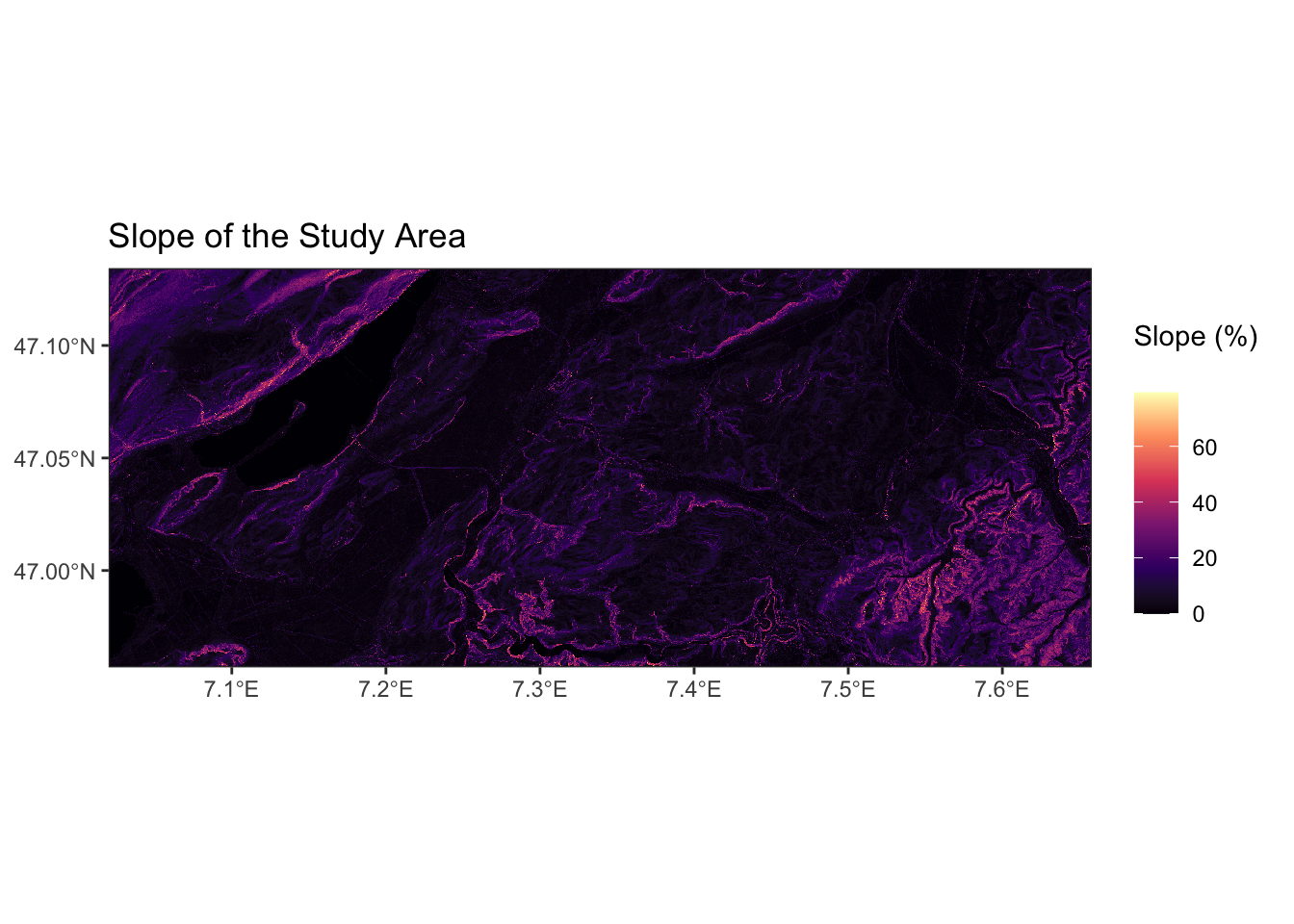
Note that the second plot has different coordinates than the upper one. That is because the data was automatically projected to the World Geodetic System (WGS84, ESPG: 4326).
This looks already interesting but we can put our data into a bit more context. For example, a larger map background would be useful to get a better orientation of our location. Also, it would be nice to see where our sampling locations are and to differentiate these locations by whether they are part of the training or testing dataset. Bringing this all together requires some more understanding of plotting maps in R. So, don’t worry if you do not understand everything in the code chunk below and enjoy the visualizations:
# To get our map working correctly, we have to ensure that all the input data
# is in the same coordinate system. Since our Bern data is in the Swiss
# coordinate system, we have to transform the sampling locations to the
# World Geodetic System first.
# To look up EPSG Codes: https://epsg.io/
# World Geodetic System 1984: 4326
# Swiss CH1903+ / LV95: 2056
# For the raster:
rasta <- project(raster_example, "+init=EPSG:4326")
# Let's make a function for transforming the sampling locations:
change_coords <- function(data, from_CRS, to_CRS) {
# Check if data input is correct
if (!all(names(data) %in% c("id", "lat", "lon"))) {
stop("Input data needs variables: id, lat, lon")
}
# Create simple feature for old CRS
sf_old_crs <- st_as_sf(data, coords = c("lon", "lat"), crs = from_CRS)
# Transform to new CRS
sf_new_crs <- st_transform(sf_old_crs, crs = to_CRS)
sf_new_crs$lat <- st_coordinates(sf_new_crs)[, "Y"]
sf_new_crs$lon <- st_coordinates(sf_new_crs)[, "X"]
sf_new_crs <- sf_new_crs |>
as_tibble() |>
dplyr::select(id, lat, lon)
# Return new CRS
return(sf_new_crs)
}
# Transform dataframes
coord_train <- df_obs |>
filter(dataset == "calibration") |>
dplyr::select(site_id_unique, x, y) |>
rename(id = site_id_unique, lon = x, lat = y) |>
change_coords(
from_CRS = 2056,
to_CRS = 4326
)
coord_test <- df_obs |>
dplyr::filter(dataset == "validation") |>
dplyr::select(site_id_unique, x, y) |>
dplyr::rename(id = site_id_unique, lon = x, lat = y) |>
change_coords(
from_CRS = 2056,
to_CRS = 4326
)# Notes:
# - This code may only work when installing the development branch of {leaflet}:
# remotes::install_github('rstudio/leaflet')
# - You might have to do library(terra) for R to find functions needed in the backend
library(terra)
# Let's get a nice color palette now for easy reference
pal <- leaflet::colorNumeric(
"magma",
terra::values(rasta),
na.color = "transparent"
)
coord_test <- df_obs |>
filter(dataset == "validation") |>
dplyr::select(site_id_unique, x, y) |>
rename(id = site_id_unique, lon = x, lat = y) |>
change_coords(
from_CRS = 2056,
to_CRS = 4326
)We now use the {leaflet} library for an interactive plot.
# Notes:
# - This code may only work when installing the development branch of {leaflet}:
# remotes::install_github('rstudio/leaflet')
# Let's get a nice color palette now for easy reference
pal <- colorNumeric(
"magma",
values(rasta),
na.color = "transparent"
)
# Next, we build a leaflet map
leaflet() |>
# As base maps, use two provided by ESRI
addProviderTiles(providers$Esri.WorldImagery, group = "World Imagery") |>
addProviderTiles(providers$Esri.WorldTopoMap, group = "World Topo") |>
# Add our raster file
addRasterImage(
rasta,
colors = pal,
opacity = 0.6,
group = "raster"
) |>
# Add markers for sampling locations
addCircleMarkers(
data = coord_train,
lng = ~lon, # Column name for x coordinates
lat = ~lat, # Column name for y coordinates
group = "training",
color = "black"
) |>
addCircleMarkers(
data = coord_test,
lng = ~lon, # Column name for x coordinates
lat = ~lat, # Column name for y coordinates
group = "validation",
color = "red"
) |>
# Add some layout and legend
addLayersControl(
baseGroups = c("World Imagery", "World Topo"),
position = "topleft",
options = layersControlOptions(collapsed = FALSE),
overlayGroups = c("raster", "training", "validation")
) |>
addLegend(
pal = pal,
values = terra::values(rasta),
title = "Slope (%)")This plotting example is based to the one shown in the AGDS 2 tutorial “Handful of Pixels” on phenology. More information on using spatial data in R can be found there in the Chapter on Geospatial data in R.
That looks great! At a first glance, it is a bit crowded but once you zoom in, you can investigate our study area quite nicely. You can check whether the slope raster file makes sense by comparing it against the base maps. Can you see how cliffs along the Aare river, hills, and even gravel quarries show high slope values. We also see that our testing dataset is randomly distributed across the area covered by the training dataset.
Now that we have played with a few visualizations, let’s get back to preparing our data. The {terra} package comes with the very useful tool to stack multiple rasters on top of each other if they share the spatial grid (extent and resolution). To do so, we just have to feed in the vector of file names list_raster:
class : SpatRaster
size : 986, 2428, 91 (nrow, ncol, nlyr)
resolution : 20, 20 (x, y)
extent : 2568140, 2616700, 1200740, 1220460 (xmin, xmax, ymin, ymax)
coord. ref. : CH1903+ / LV95
sources : NegO.tif
PosO.tif
Se_MRRTF2m.tif
... and 88 more sources
names : NegO, PosO, Se_MRRTF2m, Se_MRVBF2m, Se_NO2m_r500, Se_PO2m_r500, ...
min values : 0.8109335, 0.8742412, 0.000000, 0.000000, 0.2755551, 0.3541574, ...
max values : 1.5921584, 1.6218545, 6.965698, 7.991423, 1.6376855, 1.6430260, ... Note that above, we have stacked only a random of all available raster data (list_raster) which we have generated previously.
Now, we do not want to have the covariates’ data from all cells in the raster file. Rather, we want to reduce our stacked rasters to the x and y coordinates for which we have soil sampling data. We can do this using the terra::extract() function from the {terra} package. Then, we want to merge the two dataframes of soil data and covariates data by their coordinates. Since number of rows and the order of the covariate data is the same as the “Bern data” (soil samples), we can simply bind their columns with bind_rows():
# Extract coordinates from sampling locations
sampling_xy <- df_obs |>
dplyr::select(x, y)
# From all rasters, extract values for sampling coordinates
df_covars <- terra::extract(
all_rasters, # The raster we want to extract from
sampling_xy, # A matrix of x and y values to extract for
ID = FALSE # To not add a default ID column to the output
)
df_full <- cbind(df_obs, df_covars)
head(df_full) |>
kable()| site_id_unique | timeset | x | y | dataset | dclass | waterlog.30 | waterlog.50 | waterlog.100 | ph.0.10 | ph.10.30 | ph.30.50 | ph.50.100 | NegO | PosO | Se_MRRTF2m | Se_MRVBF2m | Se_NO2m_r500 | Se_PO2m_r500 | Se_SAR2m | Se_SCA2m | Se_TWI2m | Se_TWI2m_s15 | Se_TWI2m_s60 | Se_alti2m_std_50c | Se_conv2m | Se_curv25m | Se_curv2m | Se_curv2m_fmean_50c | Se_curv2m_fmean_5c | Se_curv2m_s60 | Se_curv2m_std_50c | Se_curv2m_std_5c | Se_curv50m | Se_curv6m | Se_curvplan25m | Se_curvplan2m | Se_curvplan2m_fmean_50c | Se_curvplan2m_fmean_5c | Se_curvplan2m_s60 | Se_curvplan2m_s7 | Se_curvplan2m_std_50c | Se_curvplan2m_std_5c | Se_curvplan50m | Se_curvprof25m | Se_curvprof2m | Se_curvprof2m_fmean_50c | Se_curvprof2m_fmean_5c | Se_curvprof2m_s60 | Se_curvprof2m_s7 | Se_curvprof2m_std_50c | Se_curvprof2m_std_5c | Se_curvprof50m | Se_diss2m_50c | Se_diss2m_5c | Se_e_aspect25m | Se_e_aspect2m | Se_e_aspect2m_5c | Se_e_aspect50m | Se_n_aspect2m | Se_n_aspect2m_50c | Se_n_aspect2m_5c | Se_n_aspect50m | Se_n_aspect6m | Se_rough2m_10c | Se_rough2m_5c | Se_rough2m_rect3c | Se_slope2m | Se_slope2m_fmean_50c | Se_slope2m_fmean_5c | Se_slope2m_s60 | Se_slope2m_s7 | Se_slope2m_std_50c | Se_slope2m_std_5c | Se_slope50m | Se_slope6m | Se_toposcale2m_r3_r50_i10s | Se_tpi_2m_50c | Se_tpi_2m_5c | Se_tri2m_altern_3c | Se_tsc10_2m | Se_vrm2m | Se_vrm2m_r10c | be_gwn25_hdist | be_gwn25_vdist | cindx10_25 | cindx50_25 | geo500h1id | geo500h3id | lgm | lsf | mrrtf25 | mrvbf25 | mt_gh_y | mt_rr_y | mt_td_y | mt_tt_y | mt_ttvar | protindx | terrTextur | tsc25_18 | tsc25_40 | vdcn25 | vszone |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4_26-In-005 | d1968_1974_ptf | 2571994 | 1203001 | validation | poor | 0 | 0 | 1 | 6.071733 | 6.227780 | 7.109235 | 7.214589 | 1.569110 | 1.534734 | 5.930607 | 6.950892 | 1.562085 | 1.548762 | 4.000910 | 16.248077 | 0.0011592 | 0.0032796 | 0.0049392 | 0.3480562 | -40.5395088 | -0.0014441 | -1.9364884 | -0.0062570 | 0.0175912 | 0.0002296 | 2.9204133 | 1.1769447 | 0.0031319 | -0.5886537 | -0.0042508 | -1.0857303 | -0.0445323 | -0.0481024 | -0.0504083 | -0.1655090 | 1.5687343 | 0.6229440 | 0.0007920 | -0.0028067 | 0.8507581 | -0.0382753 | -0.0656936 | -0.0506380 | -0.0732220 | 1.6507173 | 0.7082230 | -0.0023399 | 0.3934371 | 0.1770810 | -0.9702092 | -0.5661940 | -0.7929600 | -0.9939429 | -0.2402939 | -0.2840056 | -0.6084610 | -0.0577110 | -0.7661251 | 0.3228087 | 0.2241062 | 0.2003846 | 1.1250136 | 0.9428899 | 0.6683306 | 0.9333237 | 0.7310556 | 0.8815832 | 0.3113754 | 0.3783818 | 0.5250366 | 0 | -0.0940372 | -0.0583917 | 10.319408 | 0.4645128 | 0.0002450 | 0.000125 | 234.39087 | 1.2986320 | -10.62191 | -6.9658718 | 6 | 0 | 7 | 0.0770846 | 0.0184651 | 4.977099 | 1316.922 | 9931.120 | 58 | 98 | 183 | 0.0159717 | 0.6248673 | 0.3332805 | 1.784737 | 65.62196 | 6 |
| 4_26-In-006 | d1974_1978 | 2572149 | 1202965 | calibration | poor | 0 | 1 | 1 | 6.900000 | 6.947128 | 7.203502 | 7.700000 | 1.568917 | 1.533827 | 5.984921 | 6.984581 | 1.543384 | 1.558683 | 4.001326 | 3.357315 | 0.0139006 | 0.0070509 | 0.0067992 | 0.1484705 | 19.0945148 | -0.0190294 | 2.1377332 | 0.0021045 | 0.0221433 | 0.0000390 | 3.8783867 | 4.3162045 | -0.0171786 | 0.1278165 | -0.0119618 | -0.3522736 | -0.0501855 | -0.3270764 | -0.1004921 | -0.5133076 | 2.0736780 | 2.2502327 | -0.0073879 | 0.0070676 | -2.4900069 | -0.0522900 | -0.3492197 | -0.1005311 | -0.4981292 | 2.1899190 | 2.4300070 | 0.0097907 | 0.4014700 | 0.7360508 | 0.5683194 | -0.3505180 | 0.8753148 | 0.3406741 | 0.4917848 | -0.5732749 | 0.4801802 | -0.4550385 | 0.7722272 | 0.2730940 | 0.2489859 | 0.2376962 | 1.3587183 | 1.0895698 | 0.9857153 | 1.0231543 | 1.0398037 | 1.0152543 | 0.5357812 | 0.0645478 | 0.5793087 | 0 | -0.0014692 | 0.0180000 | 12.603136 | 0.5536283 | 0.0005389 | 0.000300 | 127.41681 | 1.7064546 | -10.87862 | -11.8201790 | 6 | 0 | 7 | 0.0860347 | 0.0544361 | 4.975796 | 1317.000 | 9931.672 | 58 | 98 | 183 | 0.0204794 | 0.7573612 | 0.3395441 | 1.832904 | 69.16074 | 6 |
| 4_26-In-012 | d1974_1978 | 2572937 | 1203693 | calibration | moderate | 0 | 1 | 1 | 6.200000 | 6.147128 | 5.603502 | 5.904355 | 1.569093 | 1.543057 | 5.953919 | 6.990917 | 1.565405 | 1.563151 | 4.000320 | 11.330072 | 0.0011398 | 0.0021498 | 0.0017847 | 0.1112066 | -9.1396294 | 0.0039732 | -0.4178924 | 0.0009509 | 0.0431735 | 0.0034232 | 0.7022317 | 0.4170935 | -0.0026431 | -0.0183221 | 0.0015183 | -0.2168447 | -0.0079620 | 0.0053904 | -0.0091239 | -0.0110896 | 0.3974485 | 0.2292406 | -0.0013561 | -0.0024548 | 0.2010477 | -0.0089129 | -0.0377831 | -0.0125471 | -0.0052359 | 0.4158890 | 0.2700820 | 0.0012870 | 0.6717541 | 0.4404107 | -0.6987815 | -0.1960597 | -0.3866692 | -0.7592779 | -0.9633239 | -0.3006475 | -0.9221049 | -0.3257418 | -0.9502072 | 0.2305476 | 0.2182523 | 0.1434273 | 0.7160403 | 0.5758902 | 0.5300468 | 0.5107915 | 0.5744110 | 0.4975456 | 0.2001768 | 0.1311051 | 0.4620202 | 0 | 0.0340407 | -0.0145804 | 7.100000 | 0.4850160 | 0.0000124 | 0.000000 | 143.41533 | 0.9372618 | 22.10210 | 0.2093917 | 6 | 0 | 7 | 0.0737963 | 3.6830916 | 4.986864 | 1315.134 | 9935.438 | 58 | 98 | 183 | 0.0048880 | 0.7978453 | 0.4455501 | 1.981526 | 63.57096 | 6 |
| 4_26-In-014 | d1974_1978 | 2573374 | 1203710 | validation | well | 0 | 0 | 0 | 6.600000 | 6.754607 | 7.200000 | 7.151129 | 1.569213 | 1.542792 | 4.856076 | 6.964162 | 1.562499 | 1.562670 | 4.000438 | 42.167496 | 0.0000000 | 0.0008454 | 0.0021042 | 0.3710849 | -0.9318936 | -0.0371234 | -0.0289909 | 0.0029348 | -0.1056513 | 0.0127788 | 1.5150748 | 0.2413423 | 0.0020990 | -0.0706228 | -0.0113604 | -0.0272214 | -0.0301961 | -0.0346193 | -0.0273140 | -0.0343277 | 0.8245047 | 0.1029889 | -0.0041158 | 0.0257630 | 0.0017695 | -0.0331309 | 0.0710320 | -0.0400928 | 0.0529446 | 0.8635767 | 0.1616543 | -0.0062147 | 0.4988544 | 0.4217250 | -0.8485889 | -0.8836724 | -0.8657616 | -0.8993938 | -0.4677161 | -0.5735765 | -0.4998477 | -0.4121092 | -0.4782534 | 0.3859352 | 0.2732429 | 0.1554769 | 0.8482135 | 0.8873205 | 0.8635756 | 0.9015982 | 0.8518201 | 0.5767300 | 0.2149791 | 0.3928713 | 0.8432562 | 0 | 0.0686932 | -0.0085602 | 8.303085 | 0.3951114 | 0.0000857 | 0.000100 | 165.80418 | 0.7653937 | -20.11569 | -7.7729993 | 6 | 0 | 7 | 0.0859686 | 0.0075817 | 5.285522 | 1315.160 | 9939.923 | 58 | 98 | 183 | 0.0064054 | 0.4829135 | 0.4483251 | 2.113142 | 64.60535 | 6 |
| 4_26-In-015 | d1968_1974_ptf | 2573553 | 1203935 | validation | moderate | 0 | 0 | 1 | 6.272715 | 6.272715 | 6.718392 | 7.269008 | 1.570359 | 1.541979 | 4.130917 | 6.945287 | 1.550528 | 1.562685 | 4.000948 | 5.479310 | 0.0054557 | 0.0043268 | 0.0045225 | 0.3907509 | 4.2692256 | 0.0378648 | 0.6409346 | 0.0022611 | -0.1020419 | 0.0161510 | 3.6032522 | 1.8169731 | 0.0346340 | 0.0476020 | 0.0378154 | 0.2968794 | -0.0179657 | -0.0137853 | -0.0146946 | 0.0060875 | 1.4667766 | 0.9816071 | 0.0337645 | -0.0000494 | -0.3440553 | -0.0202268 | 0.0882566 | -0.0308456 | 0.0929077 | 2.6904552 | 1.0218329 | -0.0008695 | 0.6999696 | 0.3944107 | -0.8918364 | -0.7795515 | -0.8864348 | -0.4249992 | 0.5919228 | 0.4304937 | 0.4614536 | 0.6559467 | 0.4574654 | 0.4330348 | 0.3299487 | 0.1889674 | 1.2301254 | 1.8937486 | 1.2098556 | 1.5986075 | 1.2745584 | 2.7759163 | 0.5375320 | 0.3582314 | 1.1426100 | 0 | 0.3005829 | 0.0061576 | 10.110727 | 0.5134069 | 0.0002062 | 0.000200 | 61.39244 | 1.0676192 | -55.12566 | -14.0670462 | 6 | 0 | 7 | 0.0650000 | 0.0007469 | 5.894688 | 1315.056 | 9942.032 | 58 | 98 | 183 | 0.0042235 | 0.6290755 | 0.3974232 | 2.080674 | 61.16533 | 6 |
| 4_26-In-016 | d1968_1974_ptf | 2573310 | 1204328 | calibration | poor | 0 | 0 | 1 | 6.272715 | 6.160700 | 5.559031 | 5.161655 | 1.569434 | 1.541606 | 2.030315 | 6.990967 | 1.563066 | 1.552568 | 4.000725 | 13.499996 | 0.0000000 | 0.0001476 | 0.0003817 | 0.1931891 | -0.1732794 | -0.1602274 | 0.0318570 | -0.0035833 | -0.1282881 | 0.0003549 | 1.5897882 | 0.8171870 | -0.0123340 | 0.0400775 | -0.0813964 | 0.0100844 | -0.0049875 | 0.0320331 | -0.0049053 | 0.0374298 | 0.7912259 | 0.3455668 | -0.0059622 | 0.0788309 | -0.0217726 | -0.0014042 | 0.1603212 | -0.0052602 | 0.0867119 | 1.0207798 | 0.6147888 | 0.0063718 | 0.3157751 | 0.5292308 | -0.8766075 | 0.8129975 | 0.5905659 | 0.1640853 | 0.5820994 | 0.6325440 | 0.8054439 | 0.7448481 | 0.6081498 | 0.3688371 | 0.2607146 | 0.1763995 | 1.0906221 | 1.0418727 | 0.8515157 | 1.2106605 | 0.8916541 | 1.2163279 | 0.4894866 | 0.2049688 | 0.7156029 | 0 | -0.0910767 | 0.0034276 | 9.574804 | 0.3864355 | 0.0001151 | 0.000525 | 310.05014 | 0.1321367 | -17.16055 | -28.0693741 | 6 | 0 | 7 | 0.0731646 | 0.0128017 | 5.938320 | 1315.000 | 9940.597 | 58 | 98 | 183 | 0.0040683 | 0.6997021 | 0.4278295 | 2.041467 | 55.78354 | 6 |
Now, not all our covariates may be continuous variables and therefore have to be encoded as factors. As an easy check, we can take the original covariates data and check for the number of unique values in each raster. If the variable is continuous, we expect that there are a lot of different values - at maximum 1052 different values because we have that many entries. So, let’s have a look and assume that variables with 10 or less different values are categorical variables.
vars_categorical <- df_covars |>
# Get number of distinct values per variable
summarise(across(everything(), ~ n_distinct(.))) |>
# Turn df into long format for easy filtering
tidyr::pivot_longer(
everything(),
names_to = "variable",
values_to = "n"
) |>
# Filter out variables with 10 or less distinct values
filter(n <= 10) |>
# Extract the names of these variables
pull("variable")
cat(
"Variables with less than 10 distinct values:",
ifelse(length(vars_categorical) == 0, "none", vars_categorical)
)Variables with less than 10 distinct values: geo500h1idNow that we have the names of the categorical values, we can mutate these columns in our data frame using the base function as.factor():
We are almost done with our data preparation, we just need to reduce it to sampling locations for which we have a decent amount of data on the covariates. Else, we blow up the model calibration with data that is not informative enough.
# Get number of rows to calculate percentages
n_rows <- nrow(df_full)
# Get number of distinct values per variable
df_full |>
summarise(across(
everything(),
~ length(.) - sum(is.na(.))
)) |>
tidyr::pivot_longer(everything(),
names_to = "variable",
values_to = "n"
) |>
mutate(perc_available = round(n / n_rows * 100)) |>
arrange(perc_available) |>
head(10) |>
knitr::kable()| variable | n | perc_available |
|---|---|---|
| ph.30.50 | 856 | 81 |
| ph.10.30 | 866 | 82 |
| ph.50.100 | 859 | 82 |
| timeset | 871 | 83 |
| ph.0.10 | 870 | 83 |
| dclass | 1006 | 96 |
| site_id_unique | 1052 | 100 |
| x | 1052 | 100 |
| y | 1052 | 100 |
| dataset | 1052 | 100 |
This looks good, we have no variable with a substantial amount of missing data. Generally, only pH measurements are lacking, which we should keep in mind when making predictions and inferences. Another great way to explore your data, is using the {visdat} package:
Alright, we see that we are not missing any data in the covariate data. Mostly sampled data, specifically pH and timeset data is missing. We also see that this missing data is mostly from the same entries, so if we keep only entries where we have pH data - which is what we are interested in here - we have a dataset with pracitally no missing data.