P-model current run
Koen Hufkens and Beni Stocker
Source:vignettes/0_p-model_current_run.Rmd
0_p-model_current_run.RmdThis workflow runs the P-model as implemented in {rsofun} with its latest version.
Calibrate
Before compiling this vignette, run
# for multiple folds
# source(here::here("analysis/01_submit_calibration_current_run.R"))
# for single fold
source(here::here("analysis/rscript_calibrate_current.R"))to calibrate parameters for five folds, split by site for spatial cross-validation. This creates five files containing calibrated parameters and the {BayesianTools} model object returned from the MCMC. Read the files.
# source(here::here("analysis/01_submit_calibration_current_run.R"))
par_calib <- readRDS(here::here("data/par_calib_light_current.rds"))| kphio | kphio_par_a | kphio_par_b | soilm_thetastar | soilm_betao | err_gpp |
|---|---|---|---|---|---|
| 0.0469513 | -0.0014005 | 21.63818 | 54.73262 | 0.4304953 | 2.249227 |
Predict and test
# load driver data for all sites
drivers <- readRDS(here::here("data/drivers.rds"))
# Combine fixed and calibrated model parameters
# NOTE: this must be consistent with values specified in calibrate_rsofun().
par_fixed <- list(
beta_unitcostratio = 146.0,
kc_jmax = 0.41,
rd_to_vcmax = 0.014,
tau_acclim = 20.0
)
par <- c(par_calib, par_fixed)
output <- runread_pmodel_f(
drivers = drivers,
par = par
) Run evaluation
This runs the eval_sofun() routine included in the
sofunCalVal package, and outputs daily, monthly and annual summary
statistics comparing observed with simulated values. We’ll retake this
same routine when running on the latest release (i.e. the workflow is
equivalent but for the release of the rsofun package)
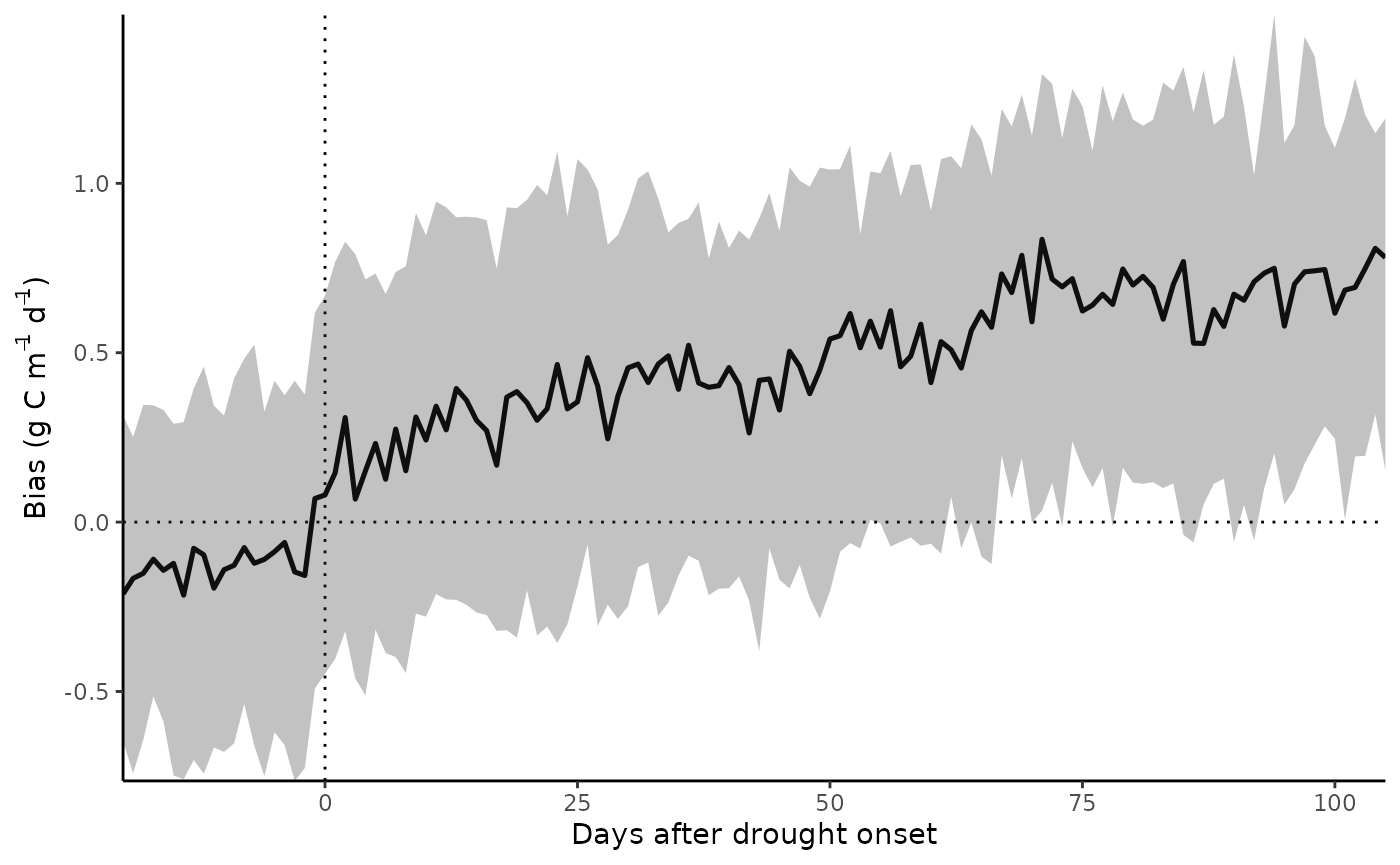
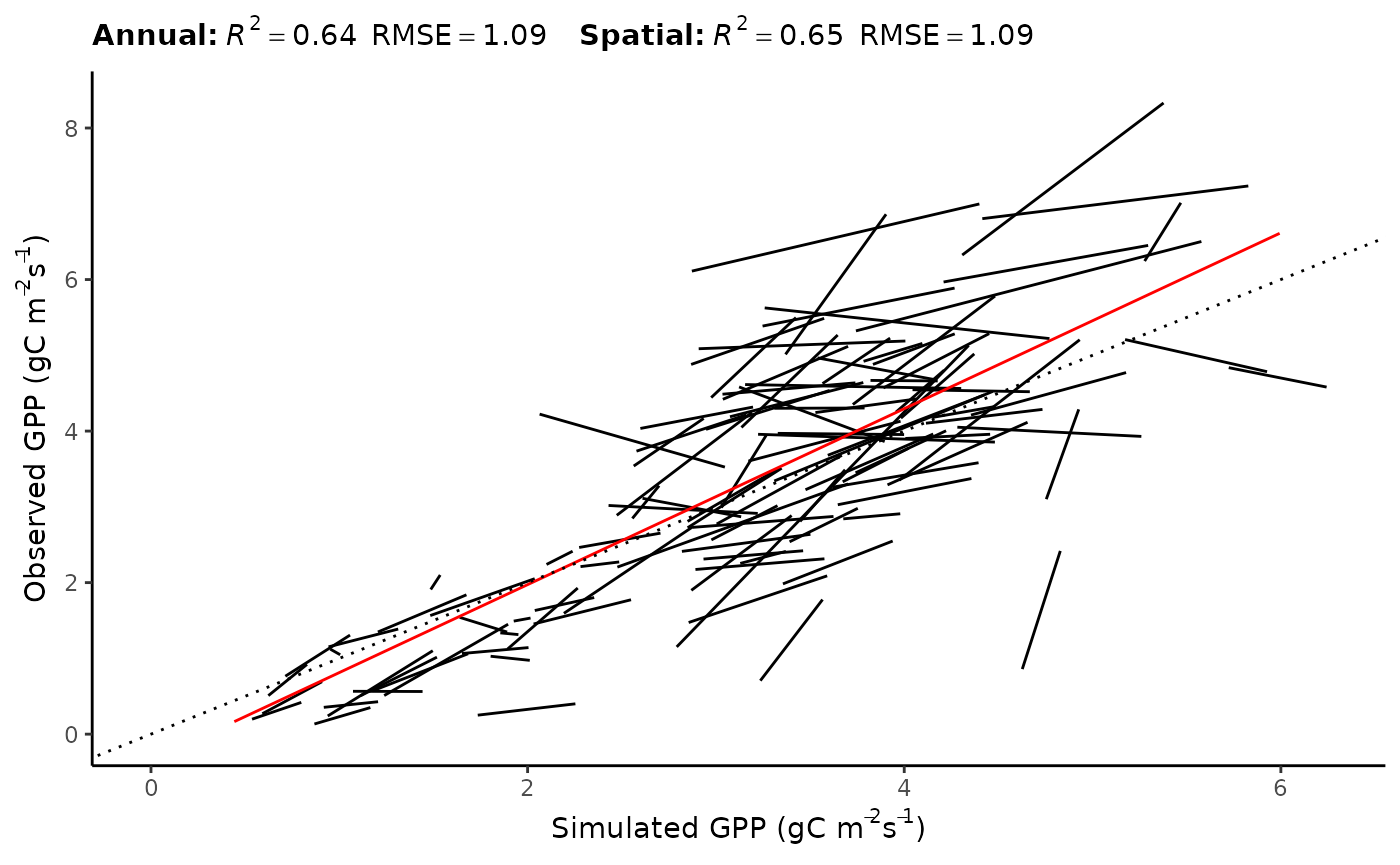
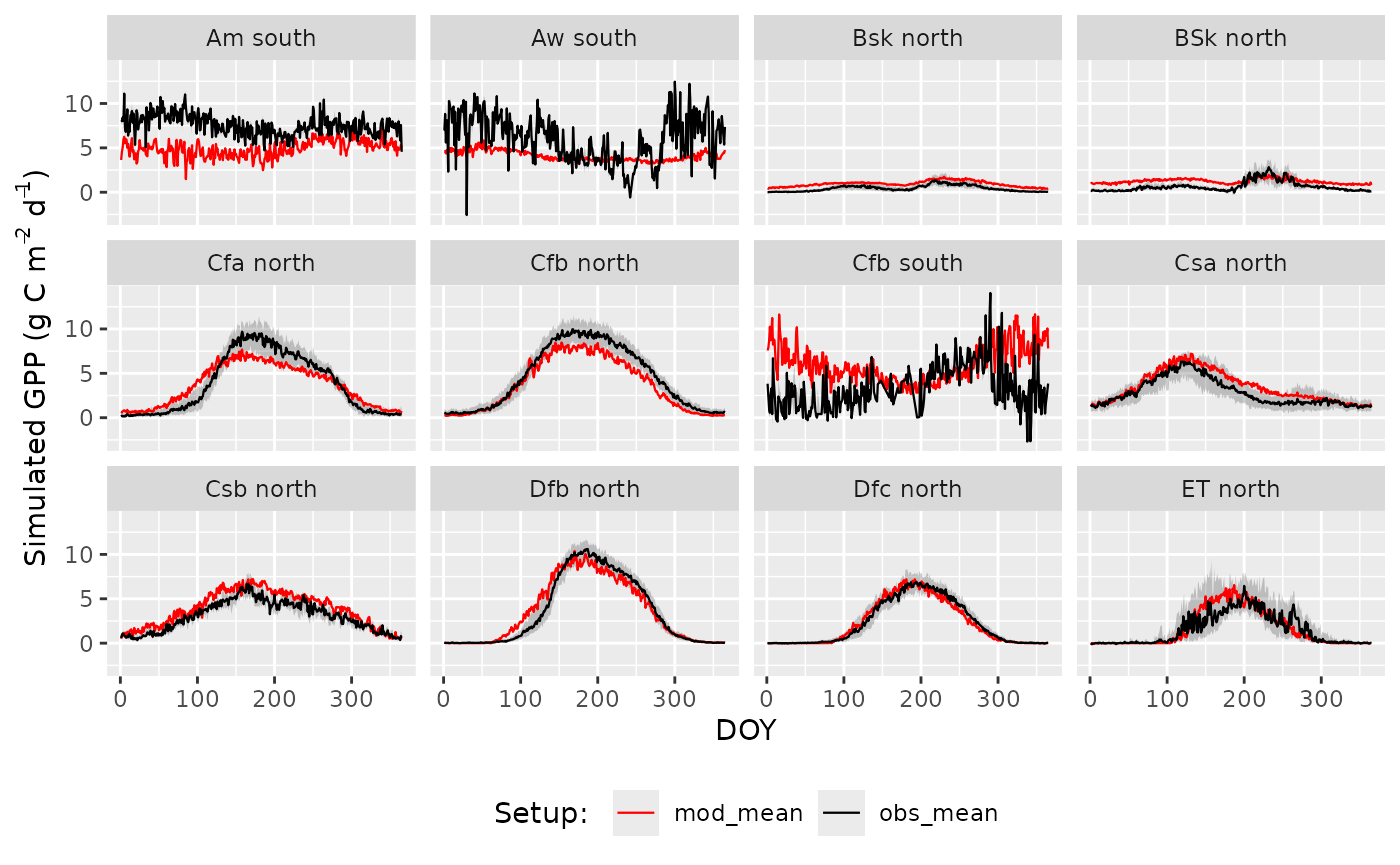
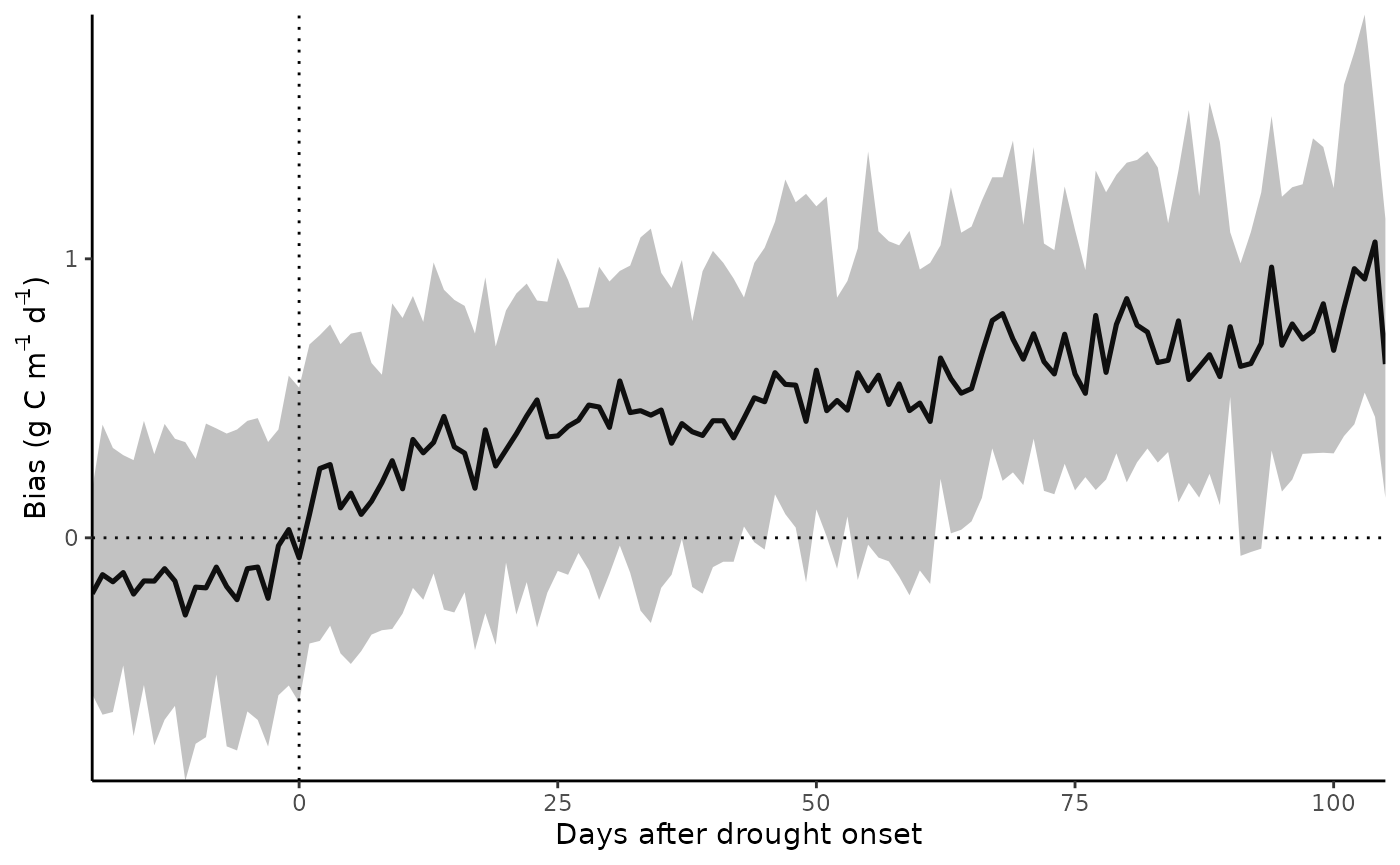
Results
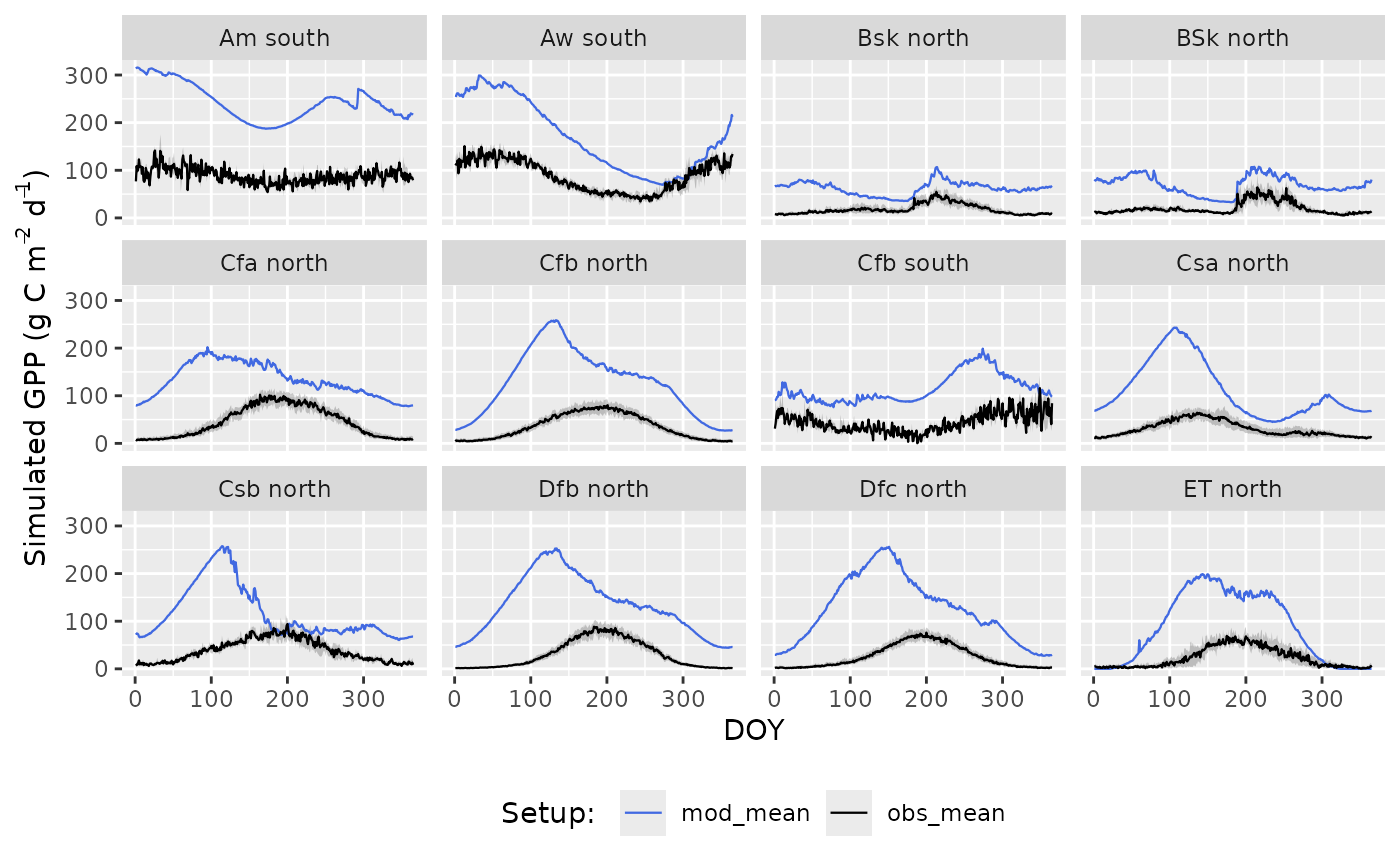
GPP
| Level | rsq | rmse | slope | bias | nvals |
|---|---|---|---|---|---|
| daily_pooled | 0.6509112 | 2.2488261 | 0.9575137 | -0.1355814 | 359373 |
| xdaily_pooled | 0.7226774 | 1.9349189 | 1.0658155 | -0.1407310 | 44981 |
| annual_pooled | 0.6398189 | 1.0898451 | 1.2085908 | -0.1778987 | 977 |
| monthly_pooled | 0.7730405 | 1.7117580 | 1.1081129 | -0.1419150 | 11664 |
| spatial | 0.6460734 | 1.0900014 | 1.1609723 | -0.1570077 | 160 |
| anomalies_annual | 0.1262824 | 0.4165609 | 0.7312536 | -0.0043831 | 977 |
| meandoy | 0.7307407 | 1.7763758 | 1.0635236 | -0.1171400 | 64539 |
| anomalies_daily | 0.2140278 | 1.5843057 | 0.4638654 | -0.0093436 | 359113 |
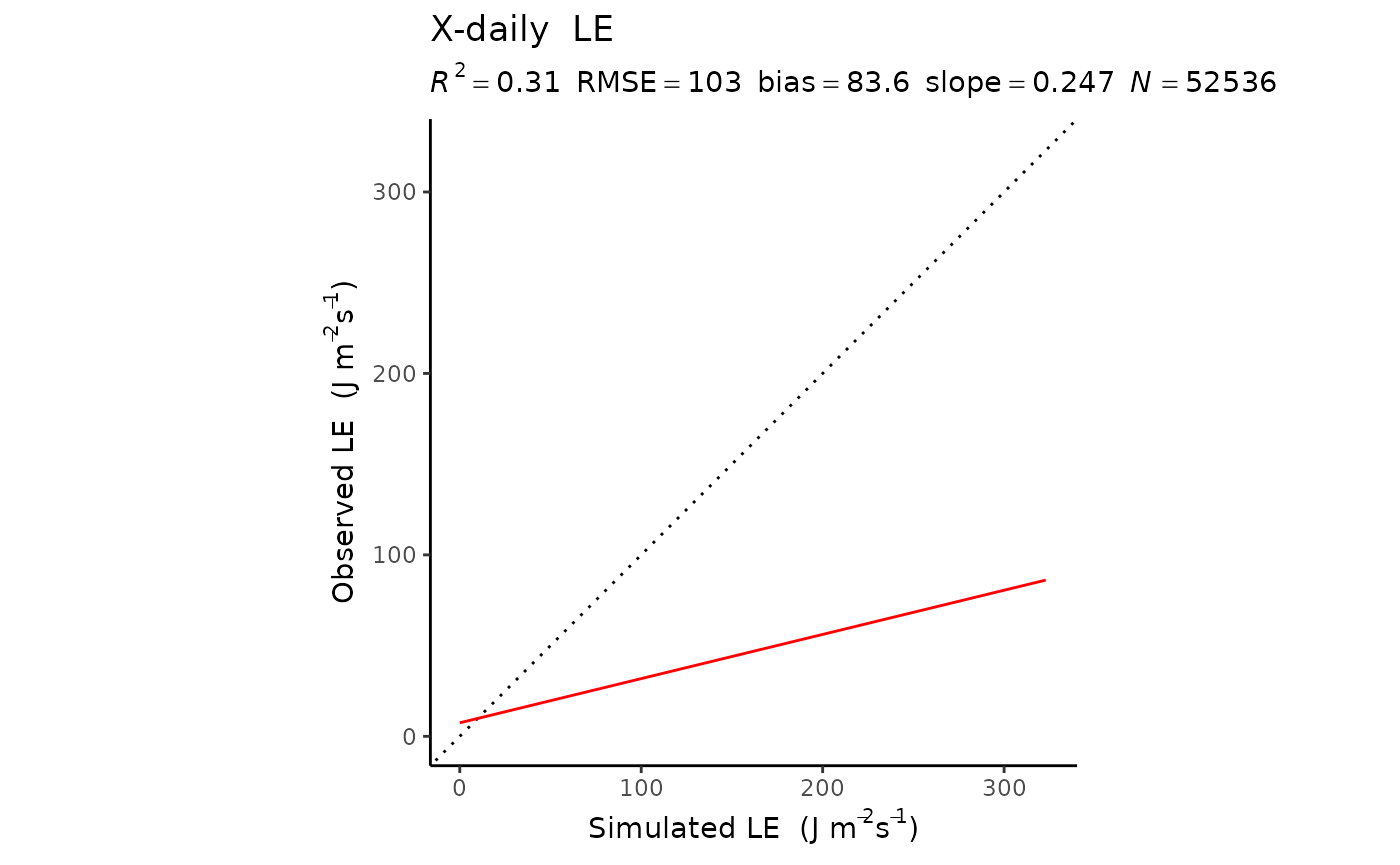
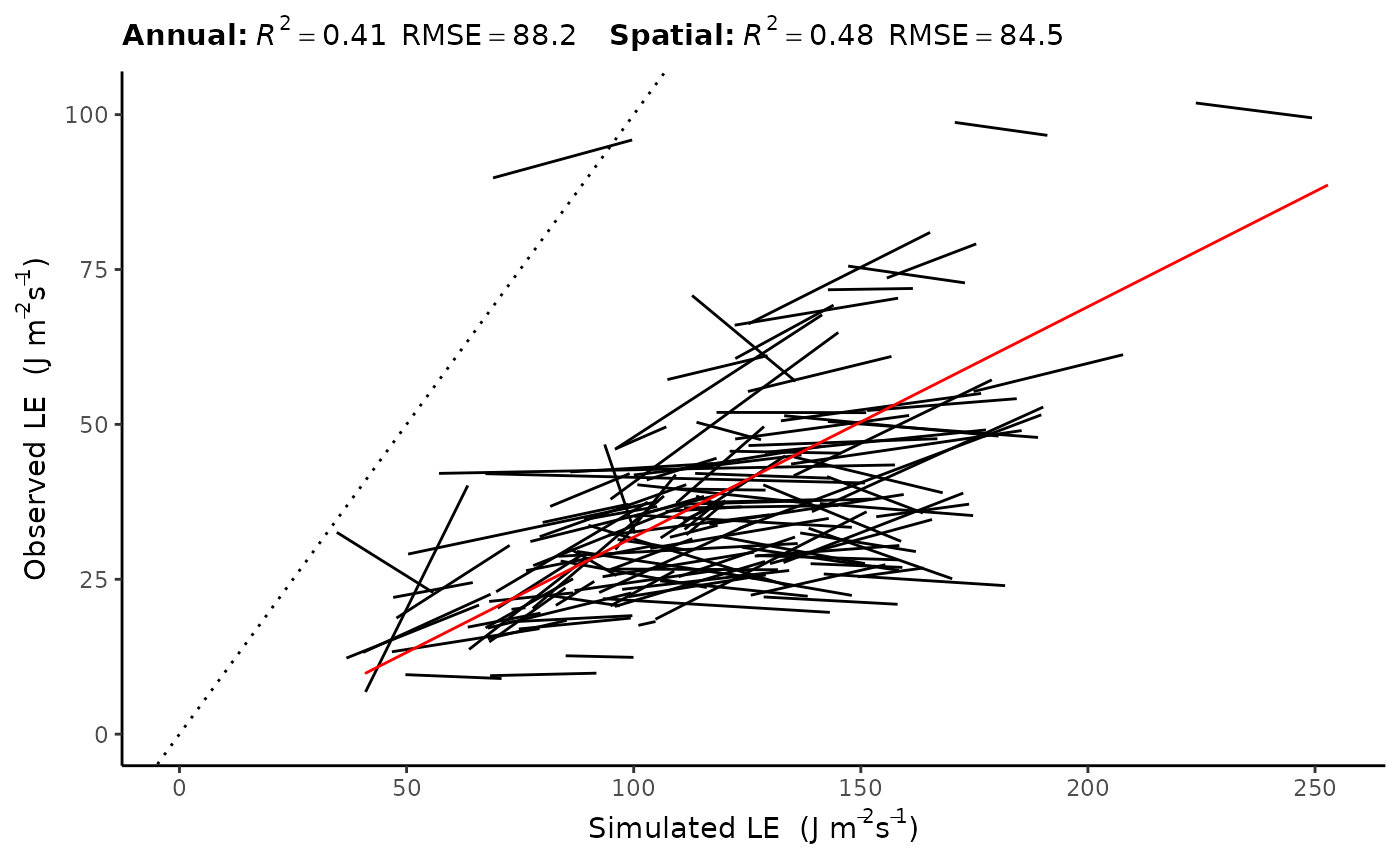
LE
| Level | rsq | rmse | slope | bias | nvals |
|---|---|---|---|---|---|
| daily_pooled | 0.2569298 | 104.77439 | 0.2455621 | 83.7466046 | 421169 |
| xdaily_pooled | 0.3051175 | 103.39162 | 0.2474844 | 83.6446622 | 52536 |
| annual_pooled | 0.4090353 | 88.17418 | 0.3098384 | 83.6653414 | 1148 |
| monthly_pooled | 0.3307161 | 102.14897 | 0.2538495 | 83.9141621 | 13579 |
| spatial | 0.4771749 | 84.47549 | 0.3719679 | 80.2417062 | 202 |
| anomalies_annual | 0.0522273 | 10.46185 | 0.1136311 | 0.0057169 | 1148 |
| meandoy | 0.3355818 | 97.26962 | 0.2848276 | 79.9400744 | 74835 |
| anomalies_daily | 0.0170511 | 29.90105 | 0.0847416 | -0.0815497 | 420863 |
Consistency with rpmodel
Since rsofun takes time series forcing, overwrite forcing with
constant values corresponding to the arguments provided to
rpmodel::rpmodel().
Setup ORG
## [1] "Are values equivalent for:"## [1] "- ci:ca: TRUE"## [1] "- GPP: Mean relative difference: 0.05018662"## [1] "- Vcmax: Mean relative difference: 0.003759071"## [1] "- Vcmax25: Mean relative difference: 0.003756366"## [1] "- Jmax: Mean relative difference: 0.003758232"## [1] "- Jmax25: Mean relative difference: 0.003756492"Appendix
## R version 4.4.3 (2025-02-28)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] here_1.0.1 ggthemes_5.1.0 knitr_1.49 lubridate_1.9.4
## [5] ggplot2_3.5.1 readr_2.1.5 tidyr_1.3.1 dplyr_1.1.4
## [9] rsofun_5.0.0.9000 rpmodel_1.2.3
##
## loaded via a namespace (and not attached):
## [1] Rdpack_2.6.2 remotes_2.5.0 rlang_1.1.5
## [4] magrittr_2.0.3 compiler_4.4.3 mgcv_1.9-1
## [7] systemfonts_1.2.1 callr_3.7.6 vctrs_0.6.5
## [10] stringr_1.5.1 profvis_0.4.0 pkgconfig_2.0.3
## [13] crayon_1.5.3 fastmap_1.2.0 ellipsis_0.3.2
## [16] labeling_0.4.3 promises_1.3.2 rmarkdown_2.29
## [19] sessioninfo_1.2.3 tzdb_0.4.0 ps_1.9.0
## [22] nloptr_2.1.1 ragg_1.3.3 purrr_1.0.4
## [25] bit_4.5.0.1 xfun_0.51 cachem_1.1.0
## [28] jsonlite_1.9.0 later_1.4.1 parallel_4.4.3
## [31] R6_2.6.1 bslib_0.9.0 stringi_1.8.4
## [34] boot_1.3-31 pkgload_1.4.0 jquerylib_0.1.4
## [37] Rcpp_1.0.14 usethis_3.1.0 DHARMa_0.4.7
## [40] httpuv_1.6.15 Matrix_1.7-2 splines_4.4.3
## [43] timechange_0.3.0 tidyselect_1.2.1 yaml_2.3.10
## [46] miniUI_0.1.1.1 curl_6.2.1 processx_3.8.6
## [49] pkgbuild_1.4.6 lattice_0.22-6 tibble_3.2.1
## [52] shiny_1.10.0 withr_3.0.2 bridgesampling_1.1-2
## [55] coda_0.19-4.1 evaluate_1.0.3 desc_1.4.3
## [58] urlchecker_1.0.1 pillar_1.10.1 reformulas_0.4.0
## [61] generics_0.1.3 vroom_1.6.5 rprojroot_2.0.4
## [64] hms_1.1.3 BayesianTools_0.1.8 munsell_0.5.1
## [67] scales_1.3.0 minqa_1.2.8 xtable_1.8-4
## [70] glue_1.8.0 GenSA_1.1.14.1 tools_4.4.3
## [73] lme4_1.1-36 fs_1.6.5 mvtnorm_1.3-3
## [76] grid_4.4.3 yardstick_1.3.2 rbibutils_2.3
## [79] devtools_2.4.5 colorspace_2.1-1 nlme_3.1-167
## [82] cli_3.6.4 textshaping_1.0.0 khroma_1.16.0
## [85] Brobdingnag_1.2-9 gtable_0.3.6 sass_0.4.9
## [88] digest_0.6.37 htmlwidgets_1.6.4 farver_2.1.2
## [91] memoise_2.0.1 htmltools_0.5.8.1 pkgdown_2.1.1
## [94] lifecycle_1.0.4 mime_0.12 bit64_4.6.0-1
## [97] MASS_7.3-64